STRATEGIC PLAN FOR SOYBEAN GENOMICS
2003
to 2007
This document chronicles the Proceedings of Soybean Genomics Research Workshop II convened in St. Louis MO on May 20-21, 2003 by:
Diane Bellis, AgSource, Inc.
H. Roger Boerma, University of Georgia
Ed Ready, United Soybean Board
Richard Wilson, USDA/ARS
TABLE OF CONTENTS
EXECUTIVE SUMMARY
INTRODUCTION
RESEARCH PRIORITIES FOR SOYBEAN GENOMICS
-DNA
MARKERS
-PLANT
GENETIC TRANSFORMATION
-GENOME
SEQUENCING AND GENE DISCOVERY
-PROTEOMICS
AND GENE FUNCTION
-BIOINFORMATICS
APPENDICES
-Process
-Participants
EXECUTIVE SUMMARY
On May 20 and 21, 2003, nineteen researchers with expert knowledge of the critical fields of soybean genomics participated in a workshop hosted by the United Soybean Board Production Committee in St. Louis, MO. The scientists reviewed the current status of soybean genomic research and reached consensus on the following strategic plan framework for outlining research priorities and significant near-term milestones that represent �quantum leaps� in the advancement of the science of soybean genomics.
Strategic Plan Framework
The structure of this plan - five strategic goals and five research objectives - provides both a program and a scientific focus to ensure that the soybean genomics community attains planned results in an effective and timely manner. The strategic goals span the programmatic range of the field of plant genomics. The research objectives address initiatives that seek to improve core capabilities in this scientific arena.
|
Strategic Goals |
Research Objectives |
|
1. Improve the utility of genetic tools for soybean genomics research |
Develop useful gene markers for soybean |
|
2. Improve the efficacy of technology for gene manipulation in genomes |
Develop useful transformation methods for functional genomic research in soybean |
|
3. Improve understanding of the organizational structure of the soybean genome |
Identify and determine the position of genes in the soybean genome |
|
4. Improve knowledge of gene identification and regulation |
Determine the biological basis for gene function, and mechanisms of gene expression Establish state-of-art interactive databases and curation mechanisms for genomic data
|
|
5. Provide bioinformatics management of genomic databases and related natural resources |
To achieve these strategic goals and scientific objectives,
this plan emphasizes achievements that hinge on teamwork throughout the soybean
genomics community. For that reason, all actions and results will be attained
in a manner that is both inclusive and open to public scrutiny. As part of this
plan, the ability of the research community to carry out and advance soybean
genomics in the U.S. public and private sectors will be evaluated in reference
to the following performance measures and expected research accomplishments in
soybean genomics over the next four years.
INTRODUCTION
Soybean�s (Glycine max L. Merr.) emergence as a global provider of vegetable oil and protein is one of the most heralded success stories of the 20th Century. Sixty years ago, soybean was grown in North America primarily as a forage crop. However, the pioneering efforts of breeders and geneticists to enhance seed production transformed soybean from a forage crop into the world�s leading source of edible protein, vegetable oil, phospholipids, dietary antioxidants (such as tocopherols and isoflavones), nutraceuticals (such as sterols), and other ingredients of foods, feeds and industrial products. Soybean researchers in the public and private sector have continued to develop improved soybean cultivars that exhibit steady genetic gains in agronomic traits such as yielding ability, disease and pest resistance, tolerance to herbicides, and seed constituent quality. These advances in plant improvement have capitalized on the inherent genetic diversity within soybean. As a result, soybean production has expanded not only in North American but also in South American and China to meet the escalating growth in global demand for soybean and soybean products. However, the competitive nature of the global soybean market has created an environment that places even greater demands on researchers to attain higher rates of progress in the improvement of this crop. Biotechnology (principally the development of soybean cultivars with genetically engineered traits) has helped meet this challenge, by providing improved soybean cultivars that allow use of more cost-effective weed control methods. Indeed, it is estimated that 80% of the U.S. commercial soybean hectarage, planted in 2003, will include a transgenic trait. Although the promise of biotechnology looms large in achieving short-term goals for soybean production, the longer-term prospects for current genetic engineering technology are clouded by apparent market-saturation and the requirement for more restrictive regulatory approvals of new innovations in transgenic cultivars. Recognizing the emerging need to address the longer-term aspects of this issue, the soybean genetics community came together in 1999 to draft a strategic plan for the future. The resultant Soybean Genomics White Paper from that historic meeting provided a road-map that was based on �quantum leap� breakthroughs in DNA-marker technology. These landmark accomplishments in SSR and SNP gene markers, made by soybean researchers, set a solid foundation for launch of a new era in soybean genetics, the characterization and interpretation of the soybean genome. These enabling technologies established concepts and provided genetic tools that have significantly advanced the study of plant genomes not only in soybean, but in all crop species.
Since publication of the Soybean Genomics White Paper, considerable progress has been made in the area of plant genomics, and soybeans in particular. For example, the soybean research community now has achieved:
� 1000 SNP markers
� a two-fold increase in efficiency of soybean transformation
� three 9000-element micro arrays for gene expression studies
� a draft physical map of the soybean genome
� a comprehensive database for soybean genomic data
In addition, the soybean genetics community joined forces with other legume crops in the creation of the U.S. Legume Crops Genomics Initiative (USLCGI). This initiative represents a landmark collaborative effort among the best minds involved in genomic research on soybean, peanut, common- and dry beans, alfalfa, peas and lentils, and model legumes. Because of dynamic advances in genomic technology, USLCGI partners have agreed to develop strategic plans for genomic research relative to their crop. In that regard, the soybean community has rejoined to assess the current status of soybean genomics, define the future direction of this science, and to update the strategic plan for soybean genomics research. Therefore, this document outlines a consensus on priority research approaches and targets for the soybean genomics community. It also establishes benchmarks for the evaluation of performance of the soybean genomics community and other organizations with interests in collaborative genomics research.
RESEARCH PRIORITIES FOR SOYBEAN
GENOMICS
DNA Markers
DNA markers are among the most versatile tools to emerge from genomics projects. They form the foundation of genetic linkage mapping and association analysis. Molecular markers are used for marker-assisted breeding, to anchor the physical map onto the genetic map, and as tools for assessing molecular variation within and between species. The current soybean molecular genetic linkage map contains 1845 markers (1017 SSRs, 745 RFLPs, and 83 other markers). Future targets for advances in soybean DNA marker technology are outlined below:
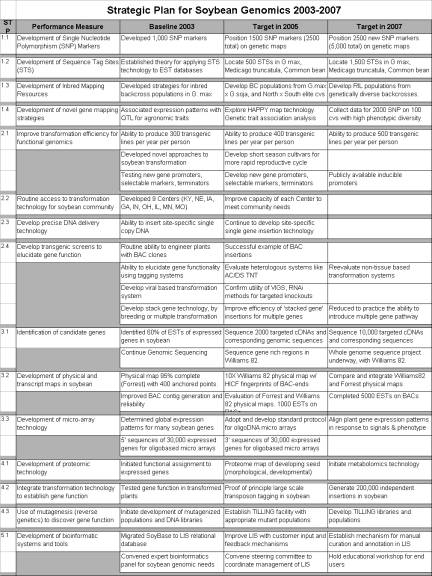
STP 1.1 Development of Single Nucleotide Polymorphism (SNP) Markers
SNPs are specific changes in DNA sequence that occur in genes as well as in intergenic regions. They can serve as biallelic genetic markers and when present in genes may alter gene function. In soybean there is approximately one SNP per 1000 bp. This translates into more than 1 million potential SNP loci in the soybean genome. SNP genetic markers are: relatively abundant, adaptable to high throughput detection, and cost effective in comparison to other DNA marker technologies. As of mid-2003 more than 3000 SNPs have been discovered in soybean. These SNPs were identified from 1000 independent DNA fragments (~ 3 SNPs/fragment) from across the genome resulting in 1000 independent SNP-tagged loci.
Goals for 2005: Discover an additional 1500 SNP containing sequence tagged sites (STS) (total of 2500) and genetically map the polymorphic SNPs in the currently available mapping populations.
Goals for 2007: Identify
an additional 2500 SNP containing STS (total of 5000) and genetically map
the polymorphic SNPs in the
currently available mapping populations.
STP 1.2 Development of Sequence Tag Sites (STS) for Cross Legume Analysis
Sequence tagged sites are specific DNA fragments that can be amplified from genomic DNA. Concomitant with the discovery of SNPs, a large number of soybean STS will be identified. Some soybean STS will be used to amplify homologous DNA fragments in other legume species. Identification of cross-species STS will enable studies of synteny across the legume family. This will facilitate the useful translation of genomic information from model species, such as Medicago truncatula, and as well as benefiting genetics and genomics research in other important crop legume species. If technically feasible, it is recommended that a mapping system in which polymorphism is not a prerequisite, such as radiation hybrids or the so-called HAPPY (HAPloid equivalents of DNA and the PolYmerase chain reaction) mapping system, be developed.
Goals for 2005: Identify 500 gene-based STS common to soybean, M. truncatula, and common bean. These STS will be used to define syntenic elements in other legume species. Explore HAPPY mapping technology.
Goals for 2007: Identify 1500 gene-based STS common to soybean, M. truncatula, and common bean. These additional STS will be used to refine syntenic associations among other legume species.
STP 1.3 Development of Inbred
Mapping Resources
Introgression lines contain single specific segments of the genome of a donor parent in a common background. For soybean the donor can be another G. max genotype, a G. soja line, or in rare cases, a related Glycine species. These lines allow the examination of donor DNA fragments in a common genetic background and the creation of useful genetic diversity. Introgression recombinant inbred lines (RIL) provide a resource for gene discovery, QTL analysis, and positional cloning. The development of a set of introgression lines requires backcrossing and extensive molecular analysis.
Goal for 2005: Develop
three backcross derived populations originating from two matings of different
G. max and G. soja parents and one mating of a northern elite cultivar and a
southern elite soybean cultivar.
Goal for 2007: Develop RIL populations from these matings with either specific G. soja-introgression fragments or distinctive genomic fragments from northern and southern cultivars that span the entire genome.
STP 1.4 Application of Association Genetics to Gene Discovery in Germplasm
Association genetics provides the opportunity to discover genes/quantitative trait loci (QTL) via direct germplasm evaluation thus bypassing the need for specially developed mapping populations. Association genetics depends upon the presence of linkage disequilibrium and relies on existing linkage between a marker(s) and a gene(s) controlling the trait of interest in an existing group of genotypes such as the germplasm lines available within the USDA Soybean Germplasm Collection. This association is detectable if one has a large number of DNA markers that are stable over evolutionary time (i.e., SNPs). Relative to a mapping population, association analysis of a diverse group of genotypes should lead to a more precise estimate of the genomic position of the gene(s) controlling the phenotypic trait of interest.
Goals for 2005: Conduct an evaluation of genetic association analysis in soybean using several select phenotypes. Initial phenotypes should include highly heritable traits such as seed protein, seed oil, and seed weight.
Goals for 2007: Identify and collect data for 2000 SNP markers on a core set of 100 genotypes that represent a substantial range in phenotypic diversity.
PLANT
GENETIC TRANSFORMATION
Soybean transformation has shown significant improvement
and enabled public and private sector production of commercial cultivars with
transgenic traits. Advances in the utility of transformation methods in soybean
have resulted from the development of selectable marker-free transgenic soybean
lines, multiple gene delivery systems, transformation and regeneration of elite
cultivars, and tissue-specific and inducible promoters. However, there is a
need for additional advances in transformation efficiency for applications in
functional genomics. Use of this technology in the discovery of gene function
is expected to grow to the point that by 2005 research progress may be impeded
by insufficient capacity of facilities to provide this service. Therefore, to
meet pending demand for this technology, improvements are needed in areas that
help ensure greater efficiency and effectiveness of soybean
transformation.
STP 2.1 Improve the Efficiency of Transformation for Functional Genomics
The development of novel approaches will be based on a better understanding of the factors that influence induction and regeneration of soybean tissue cultures. In addition, testing of new gene promoters, selectable markers, and gene coding terminators can lead to increases in transformation rates. The availability of tissue-specific gene promoters will increase the range of traits that can be improved by genetic engineering. A main limitation to high throughput functional genomics of soybean is the long growth period and large size of the plant. Development of a short life-cycle soybean genotype as an experimental system should be considered. Viral induced gene silencing (VIGS) systems, especially in somatic embryo cultures, and other rapid gene discovery methods for soybean will be very useful.
Goals for 2005: Ability to produce 400 transgenic lines
per year per person. Develop
non-tissue culture based transformation systems. Develop and test new gene promoters, selectable markers, and
terminators.
Goals for 2007: Ability to produce 500 transgenic lines
per year per person. Develop a
short season genotype for more rapid reproductive cycle. Have inducible promoters publicly
available.
STP
2.2 Routine Access to Transformation Technology for the Soybean Community
Success in improving soybean transformation and the need for high throughput technologies has created the need for the establishment and coordination of plant-growth and stock-center capacity to characterize, maintain, and distribute the developed stocks. Coordination and distribution of materials and skill between transformation laboratories will help accelerate the transfer of transformation technology to other laboratories. Support and infrastructure to maintain, characterize, and distribute seeds is essential.
Goal for 2005: Improve capacity of each Center to meet community needs.
STP 2.3 Technology to Deliver DNA Precisely
Precise integration technology should be developed for a wide range of transformation methods, including direct DNA delivery and Agrobacterium-based transformation. The current transformation methods deliver DNA randomly and imprecisely into the soybean genome. Due to these unpredictable insertion patterns, many transgenic plants do not express the phenotype in a predictable and stable manner. In addition, it would be useful to remove selectable markers and other �carry-along� DNA after they are no longer needed. The goal of this research is to develop technology for delivering DNA precisely into the soybean genome. This precision will include site-specific insertion, single copy insertion, and �clean delivery� (i.e., removal of unneeded �carry-along� DNA). This technology will furthermore lead to the precise integration of long DNA inserts, such as a bacterial artificial chromosome (BAC) clones, and the opportunity to introduce multiple genes into the same location (i.e., directed �gene stacking�). In the future, the site-specific integration process will enhance the frequency of stable DNA incorporation. For some types of genes, their function can only be definitively confirmed by testing the cloned genes in a soybean plant. Strategies to elucidate gene functionality in plant systems that should be pursued include: ability to routinely engineer plants with bacterial artificial chromosome (BAC) clones; ability to elucidate gene functionality by using transposon tagging systems to disrupt genes and study the subsequent up- and down-regulation of gene expression; and development of a viral-based transformation system, which would allow the screening of large numbers of genes via their transient expression.
Goal for 2005: Continue to develop site-specific
single gene insertion technology.
STP 2.4 Develop Transgenic Screens to Elucidate Gene Function
New technologies based on insertional mutagenesis using a range of transposon tagging strategies and targeted RNAi approaches are being developed. Continuing to improve the efficiency of extant systems will enhance these efforts. To accelerate applications of functional genomic approaches, a non-tissue culture transformation method for soybean is desirable.
Goals for 2005: Evaluate heterologous transposon
systems such as Ac/Ds and Tnt1.
Evaluate and confirm utility of VIGS/RNAi methods in somatic embryo and
whole plant approaches for targeted knockouts. Reduce to practice the ability to introduce a multiple-gene
pathway. Achieve a successful
example of BAC insertions.
GENOME SEQUENCING and GENE DISCOVERY
The genome of the soybean is approximately 1 x 109 bp and is estimated to contain 50,000 to 100,000 genes. These genes are responsible for all the pathways and functions of growth and development. The identification of candidate genes is critical for robust application of marker assisted selection, comparative analyses between genomes, and the process of understanding their function. An association with phenotype is essential to understanding how plants have adapted to the environment and how they ultimately affect plant productivity and health.
STP 3.1 Discover Soybean Genes
Gene discovery is a primary research priority in the field of genomics. It is the foundation of all functional analyses and is the ultimate target of most structural and physical genetic analyses. More than 300,000 partial gene sequences have been obtained from expressed soybean genes (expressed sequence tags; ESTs). Although this type of information provides crucial information on gene identity and gene evolution it is often necessary to have the entire expressed gene sequence in order to take full advantage of genomic tools for marker development. In order to gain information on introns as well as flanking genomic DNAs (important for understanding of gene regulations, but also important for marker development) it is necessary to obtain corresponding genomic sequence for the expressed gene.
Many of the goals of soybean genomics come ultimately from knowledge of the genome sequence. The July 2001 U.S. Legume Crops Genomics Workshop White Paper (http://www.legumes.org/) cites the sequencing of the gene-rich regions of soybean (estimated at ~340 Mb) as one of its top priorities. This is an important, achievable priority. This resource will be made more valuable by additional efforts to anchor this sequence to the physical and genetic maps. The ultimate goal should be the eventual sequencing of the entire soybean genome. Currently, this may be impractical due to the large size and concomitant expense of sequencing. However, as sequencing costs decline, achieving the complete soybean genome sequence will become an achievable goal. More immediate efforts to obtain the sequence of the gene-rich regions will aid this process and speed its completion.
Goals for 2005:
Sequence 2,000 full-length cDNAs and corresponding genomic
sequences. Have in place, in
Williams 82, a targeted genome sequencing project focusing on gene-rich
regions.
Goals for 2007: Sequence 10,000 full-length cDNAs and corresponding genomic sequences. Have in place, in Williams 82, a whole-genome sequencing project.
STP 3.2 Create Physical and
Transcript Maps of Soybean
Genome sequencing is a quantum-leap technology much like Watson and Crick�s discovery of the structure of DNA. Gene localization, which is ideally based on a fully sequenced genome, includes the creation of a physical map anchored with genetically mapped gene sequences. This is the starting point for localizing and cloning genes and sequencing the soybean genome. A complete physical map requires that a BAC library contains a minimum tile of clones for the genotype to be whole-genome sequenced (Williams 82 and/or another selected cultivar). These BACs will be fingerprinted, assembled into contigs, and their BAC-ends sequenced. Completion of a physical map with BAC-end sequences will help accomplish several other stated goals such as SNP development, genetic anchoring of physical maps, sequencing of gene rich regions, whole genome sequencing, and help to reveal ancient duplications within the soybean genome.
An integrated soybean genome map would increase the efficiency of crop improvement through application in functional genomics, maker assisted breeding, and transformation. This map is also critical to advancing numerous genomic goals such as targeted sequencing, candidate gene identification, and comparative mapping. The goal is to create a 95% complete physical map of the soybean genome encompassing a complete tile path from Williams 82; the same genotype for which a large EST resource exists. In order to assist in contig assembly and to create STS for each BAC, the ends of BACs used in the contig assembly will be sequenced. Before initiating a Williams 82 physical map, an evaluation of the efficiency of methods and the synergies for resolving duplicated, homoeologous regions will be completed. The utility of Medicago truncatula sequences for soybean map resolution will be determined. At the initial assembly of the Williams 82 physical map, it is recommended that the physical maps of Forrest and Williams 82 be independently peer reviewed. An additional research area is the establishment of a transcript map anchored to the physical and genetic maps.
Goals for 2005:
Generate a more complete and accurate physical map incorporating more Williams
82 and other cultivar clones.
Generate BAC-end sequences on sufficient Williams 82 BACs used in the
construction of the physical map to constitute a 10 X coverage of the genome. Conduct an independent evaluation of
the Forrest and Williams 82 physical maps. Place 1,000 ESTs on BACs in the physical map.
Goals for 2007: Compare and integrate Williams 82 and
Forrest physical maps. Complete
the placement of 5,000 ESTs on BACs in the physical map.
STP 3.3 Determine Soybean Gene
Expression with Advanced Micro-array Technology
All traits of living organisms are the consequence of gene expression. Information contained in the genes is translated into products that direct life functions. An understanding of the mechanisms regulating the genes that control important crop traits is a prerequisite to manipulating them to advantage.
Most important traits are specified by members of small gene families. Often closely related members of these gene families are differentially expressed at different development times and places. For this reason �paralogue-specific� technologies must be developed and applied. In addition, most traits are the result of complex interactions among numerous genes. For this reason, universal gene-expression technologies must be developed and applied.
The purpose of assigning function is to discover the genes
of agronomic importance. The
assignment of function to genes and the development of `paralogue-specific'
microarrays proceed at several levels. First it is necessary to have a nearly
full-length cDNA sequence that includes sequence at the 3' end of the gene.
There are already approximately 27,000 3' sequences derived from `unigenes' in
soybean. To represent the population of soybean expressed genes more fully, it
is recommended that 3' sequence be obtained from an additional 30,000 unigene
cDNAs identified from the Public Soybean EST collection. Once this is
accomplished, necessary arrangements should be made to provide to the soybean
community access to microarrays comprised of oligonucleotides representing
30,000 to 50,000 distinct soybean genes. This large number of represented genes
will be necessary to determine the expression patterns of genes in tissues and
organ systems of the plant by measuring the expression of thousands of genes at
a time (i.e., �global� expression patterns). Expression comparisons under conditions including pathogen
challenge, symbiont infection, heat, cold, drought stresses, and nutrient
limitations will yield classes of genes involved in these critical
processes. Expression profiles of
many agronomically important genotypes containing traits of economic importance
and QTL may also aid in assigning function. Expression profiling will yield the information needed to
select promoters useful for plant transformation.
Goals for 2005:
Migrate microarray technologies to oligo-based microarrays. Generate 3' sequences of an additional
30,000 soybean unigenes.
Goals for 2007: Characterize plant gene expression
patterns in soybean in response to abiotic and biotic signals.
PROTEOMICS and GENE FUNCTION
Genes encode proteins, and proteins carry out enzymatic functions. Important phenotypes in soybean (yield, oil, and protein content in seeds) are determined by gene function. Therefore to improve agronomic traits, the function of genes must be manipulated. Before this can be achieved, the function of each gene in the genome must be identified. Although DNA microarrays measure mRNA expression at the genomic level, results from this method do not always reflect the amount of protein that is derived from expression of a gene. Because proteins frequently specify the phenotype, determining the amount of specific proteins is important. Classically, gene function has been addressed by detailed biochemistry on single gene products (enzymes). However, the information required for genome-wide analysis makes this approach impractical. Therefore, a genome wide approach is required to determine gene function.
STP 4.1 Develop Proteomic
Technology to Determine Gene Function
Proteomics is a technology that relies on quantitative mass spectrometry to identify gene products and is based on matching the masses of tryptic digest fragments to a database of known proteins. More recently, the term proteomics has been applied to any approach that measures protein function at a genomic level. For instance, researchers can now apply methods to identify protein-protein interactions in a cell. Many proteins act in multi-protein complexes. Understanding these associations will help to better define protein function. It is recommended that a detailed proteomic analysis of the regulation of protein and oil synthesis be initiated in developing seed because of the importance of these constituents to the value of soybean as a commodity.
Goals for 2005:
Develop a proteome map of developing seed.
Goals for 2007:
Initiate metabolomics technology.
STP 4.2 Application of
Transformation Technology to Determine Gene Function
Geneticists have typically addressed gene function through mutation, and have deduced gene function based on an observation of the mutant phenotype. With the advent of efficient soybean transformation, this classical method can be applied at the genomic level by transposon-induced mutations. Two systems, Ac/Ds (from maize) and the retro-transposon Tnt1 (from tobacco), are being developed. These systems should enable broad-range deletion of genes (gene-knockouts) using transposon tagging in soybean to help determine gene function.
Goals for 2005:
Demonstrate proof of principle of large-scale transposon tagging in
soybean.
Goals for 2007:
Generate 200,000 independent insertions in soybean.
STP 4.3 Application of Reverse
Genetics to Determine Gene Function
Targeted induced local lesions in genes (TILLING) is a complimentary, high-throughput mutation based system. It is a PCR-based, reverse genetics method that permits the identification of point mutations in pre-selected genes. Given a sufficiently large, highly mutated soybean population, point mutations in any gene can be identified. Because of the long-term importance in the functional assignment of genes, it is recommended that TILLING populations and libraries should be developed as a public genetic resource. In addition, a TILLING facility should be established to coordinate use of this technology for the determination of gene function and to supply germplasm with specific mutations to breeding programs.
Goals for 2005:
Establish a common use TILLING facility with appropriate mutant
populations.
Goals for 2007: Develop TILLING libraries and
populations as a new genetic resource.
BIOINFORMATICS
Genomics projects are currently underway for several model legumes as well as for soybean and other crop legumes. These projects are resulting in the collection, storage, and analysis of many data points (i.e., sequences, expression levels, map positions). Collecting, storing, manipulating, analyzing and retrieving this vast amount of information require radically different techniques and technologies than previously used in biological studies. Further, this disparate collection of data needs to be interlinked based on a logical mapping of biological data types to one another. Researchers must be able to traverse the data from QTL to their relative locations on physical maps and, ultimately to sequence maps containing corresponding genes. Genes must be related to gene products that can be associated with biochemical pathways, allowing researchers to discover the molecular basis for phenotypic traits. Informatics components can be separated into the development of infrastructure and tools and the application of those tools to synthesize information into useable results. Infrastructure needs include the development of relational database management systems, visualization tools, algorithm development, distributed computing, storage systems, and networking. Information integration is a biological problem, which includes pathway reconstructions, understanding of developmental processes, and inferring likely phenotypic information.
The Legume Information System (LIS) was conceived to be a comparative legume resource, populated initially with data from G. max, M. truncatula and Lotus japonicus. A major bioinformatics goal is to develop a robust means of comparative transcript analysis, initially between the G. max, M. truncatula and L japonicus, and eventually including unigenes from Arabidopsis thaliana as a non-legume species. The results of comparative transcript analysis are bins of sequences based on origin. This data will be immediately useful to legume researchers who are interested in soybean-specific expression, for example. This is also the first step towards leveraging model plants to gain insights into crop species.
The second step in comparative analysis planned for LIS involve decorating genomic sequence data with the shared consensus generated as above. Currently, the genomic component of LIS uses consensus sequences generated by the transcript component of LIS for each species, of which there are currently over 150,000. Mapped gene sequences help identify gene-rich regions, help validate or refute gene models and provide data to help build scaffolding to bridge the genomic-physical map-linkage map gulf. Structural information about genomic regions may also shed light on gene families and certainly helps to address evolutionary questions concerning species relatedness. Analyzing gene structure in a genomic context is a powerful comparative genomic tool enabling identification of regions of micro- and macro- synteny.
STP 5.1 Development of Bioinformatic Systems and Tools
Starting in 2003, map data (linkage and physical) and associated metadata (authors, affiliations, literature etc.) will be ported from SoyBase into the relational CMAP database and visualization software developed by Ken Clark at Cold Spring Harbor. CMAP will be modified to interoperate seamlessly with the LIS and will feature automated linkage of sequence-based markers to EST and genomic data housed in LIS. Beginning in 2004, biochemical pathway data will also be ported from SoyBase into LIS as well as pathology, transformation data, and the other remaining data classes with the goal of completely subsuming SoyBase by 2005.During this transition, mechanisms for manual curation of soybean data will be developed, evaluated, and implemented.
The usefulness of genomic databases is partially the result of the middle-ware and the underlying engine. The ability of the user to comprehend the databases capabilities and to maneuver through the various levels of data is also critical. To facilitate the ease by which data can be viewed, manipulated, and retrieved from LIS, major improvements will be made to the existing LIS user interface, including development of a novel, graphic-based query interface which will facilitate data browsing and exploration.
A Steering Committee of legume researchers and
bioinformaticists to guide the development of LIS will be convened. As the
development of LIS progresses, it will be critical to incorporate ideas and
suggestions from the legume community. To accomplish this LIS will solicit
input on the perceived needs of the legume research community, which will
directly influence the system�s design and user interface development. This
will be accomplished by periodically convening panels of users to participate
in workshops and by providing a forum for online comments.
Goals for 2005:
Improve LIS by adding data generated through new technologies and by
incorporating input from legume researchers. Convene a Steering Committee to coordinate the management of
LIS. Establish a mechanism for
manual curation and annotation.
Migrate mapping and biochemical pathway components of SoyBase into the
LIS relational structure.
Goals for 2007:
Complete LIS as a relational database and knowledge discovery tool.
Incorporate new data types and functionality as determined by user panels and
the Steering Committee.
APPENDIX - Process
On May 20 and 21, 2003 nineteen expert researchers with knowledge in structural and functional genomics, plant transformation, and bioinformatics participated in a workshop hosted by the United Soybean Board Production Committee. Over the course of the two days, the scientists reviewed the current status of soybean genomic resources and discussed alternative approaches to efficiently acquire the additional information required to unravel the genetic information contained within this critical legume crop. The group achieved consensus on the most important soybean genomics priorities for the next five years. These priorities are reported in the Strategic Plan for Soybean Genomics 2003 to 2007.
The workshop was planned by: Dr. Diane Bellis of AgSource, Inc., a subcontractor with the United Soybean Board focusing on Federal Research Coordination; Dr. Roger Boerma, Distinguished Research Professor and Director of the University of Georgia Center for Soybean Improvement; Dr. Ed Ready, Production Program Manager for the United Soybean Board; and Dr. Richard Wilson, National Program Leader for the Agricultural Research Service in Oilseeds and Bioscience. Ed Ready facilitated the workshop. We wish to acknowledge Dr. Randy Shoemaker�s assistance in the preparation of the initial draft of the strategic plan.
The organizers wish to thank Ms. Lisa Childs for the local
meeting arrangements and her assistance during the workshop. The guidance, observations, and input
of Bryan Hieser, and Jim Sallstrom, USB Directors, Marc Curtis, American
Soybean Association, Lyle Roberts, Illinois Soybean Promotion Board, and Keith
Smith, representing the Iowa Soybean Promotion Board were critical in the
development and completion of this plan.
APPENDIX - Participants
T.E. (Tom) Clemente
Department of Agronomy
N308 Beadle Center
University of Nebraska
Lincoln, NE 68588
Tel: 402-472-1428
FAX: 402-472-3139
E-mail: tclemente1@unl.edu
Timothy W. Conner
Director, Global Oilseeds Technology
Monsanto Company, BB1E
700 N. Chesterfield Pkwy
Chesterfield, MO 63017-1700
Tel: 636/737-6007
FAX: 636/737-7507
Cell: 314 308-7303
E-mail: timothy.w.conner@monsanto.com
Perry B. Cregan
Soybean Genomics and Improvement Laboratory
USDA-ARS
Bldg. 006, Room 100
Beltsville, MD 20705
Tel: (301) 504-5070
FAX: (301) 504-5728
E-mail: creganp@ba.ars.usda.gov
Randy Dinkins
University of Kentucky
Department of Agronomy
105 Veterans Road, Room 311
Lexington, KY 40546
Tel: 859-257-5020
FAX: 859-257-7125
E-mail: rddink1@uky.edu
David Grant
G304 Agronomy Hall
Iowa State University
Ames, IA 50011
Tel: 515/294-1205
FAX: 515/294-2299
E-mail: David.Grant@ars.usda.gov
Scott Jackson
Purdue University
915 W. State Street
Department of Agronomy
West Lafayette, IN 47907
Tel: 765/496-3621
FAX: 765-496-7255
E-mail: sjackson@purdue.edu
David A. Lightfoot
Dept. of Plant, Soil, and General Agriculture
Southern Illinois University
Genomics Core Facility, PSGA, SUIC, MC4415
Carbondale,
IL 62901-4415
Tel: 618/453-1797
FAX: 618/453-7457
E-mail: ga4082@siu.edu
Ted Klein
Pioneer/DuPont Crop Genetics
Stine-Haskell Research Center
Building 614
Newark, DE 19714-0030
Telephone: (302) 283-2403
Fax: (302) 283-2417
E-mail: Ted.m.klein@Pioneer.com
Niels C. Nielsen
Purdue University/USDA-ARS
915 W. State Street
Agronomy Department
West Lafayette, IN 47907-1150
Tel: 765/494-8057
FAX: 765/494-6508
E-mail: nnielsen@purdue.edu
Henry T. Nguyen
Department of Agronomy, Plant Sciences Unit
1-87 Agriculture Building
University of Missouri � Columbia
Columbia,
MO 65211
Tel: 573/882-5494
FAX: 573/882-1469
E-mail: nguyenhenry@missouri.edu
Brian Scheffler
USDA-ARS MSA Genomics Facility
141 Experiment Station Road
Stoneville, MS 38776
Tel: 662-686-5454
FAX: 662-686-5372
E-mail: bscheffler@ars.usda.gov
Randy C. Shoemaker
G401 Agronomy Hall
Iowa State University
Ames, IA 50011
Tel: 515/294-6233
FAX: 515/294-2299
E-mail: rcsshoe@iastate.edu
David Somers
Agronomy and Plant Genetics
University of Minnesota
1991 Upper Buford Ave.
St Paul, MN 55113
Tel: 612/625-5769
FAX: 612/625-1268
E-mail: somers@biosci.cbs.umn.edu
James E. Specht
Dep. of Agronomy & Horticulture
322 Keim Hall, East Campus
University of Nebraska
Lincoln, NE 68583-0915
Tel: 402/472-1536
FAX: 402/472-7904
E-mail: jspecht1@unl.edu
Gary Stacey
University of Missouri
Department of Plant Microbiology and Pathology
108 Waters Hall
Columbia, MO 65211
Tel: 573-884-4752
FAX 573-882-0588
E-mail: staceyg@missouri.edu
Lila Vodkin
University of Illinois
384 ERML, Department of Crop Sciences
Champaign, IL 61821
Tel: 217/244-6147
FAX: 217/333-4582
E-mail: l-vodkin@uiuc.edu
Mark Vaudin
Monsanto
800 North Lindbergh Blvd.
N2NA
St. Louis, MO 63167
Tel: 314/694-3866
FAX: 314-705-8084
E-mail: mark.vaudin@monsanto.com
Mark Waugh
NCGR
2935 Rodeo Park Drive East
Santa Fe,
NM 8755
Tel:
505-995-4446
FAX:
505-995-4432
E-mail: mew@ncgr.org
Nevin
Dale Young
495 Borlaug Hall
University of Minnesota
St. Paul, MN 55108
Tel: 612/625-2225
FAX: 612/625-9728
E-mail: neviny@umn.edu
Bryan Hieser
United Soybean Board
5515 Mackinaw Road
Minier, IL 61759
Tel: (309) 392-2537
FAX: (309) 392-2890
E-mail: hieserb@trianglenet.net
Jim Sallstrom
United Soybean Board
58552 276th Street
Winthrop, MN 55396
Tel: (507) 647-2546
FAX: (507) 647-4444
E-mail: jimsall@means.net
Marc
Curtis
American Soybean Association
PO Box 958
105 California Ave
Leland, MS 38756
Tel: (662) 686-2321
FAX: (662) 686-2321
E-mail: mscfarm@tecinfo.com
Lyle Roberts
Illinois Soybean Board
1605 Commerce Parkway
Bloomington, IL 61704
Tel: (309) 663-7692
FAX: (309) 663-6981
E-mail: robertsl@ilsoy.org
Keith Smith
Keith Smith & Associates
15 Winchester Road
Farmington, MO 63640
Tel: 573-756-2284
FAX: (573) 756-2821
E-mail: ksmith@i1.net
Workshop Organizers
|
Diane Bellis AgSource, Inc. 600 Pennsylvania Ave SE Washington, DC 20003 Tel: 304/275-3087 Cell: 202/412-0582 E-mail: dbellis@agsourceinc.com |
H. Roger Boerma 111 Riverbend Road Center for Applied Genetic Technologies |
|
Ed Ready 540 Maryville Centre Drive Ste LL5 St. Louis, MO 63141 Tel: 314/579-1598 FAX: 314/579-1599 E-mail:
ed_ready@sba.com |
Rich Wilson USDA/ARS/NPS 5601 Sunnyside Avenue Rm 4-2214 Beltsville, MD 20705 Tel: 301/504-4670 E-mail: rfw@ars.usda.gov |